New method improves efficiency of shRNA screening
Mon, Jun 5, 2017-
Tags
A group of researchers came up with a new method that improves the efficiency of short-hairpin RNA (shRNA) screening, which identifies biochemical interactions that produce pharmacological effects (known as mechanisms of action). Using this method, they found the gene that regulates cytotoxicity by aurilide B, a natural marine product that induces apoptosis, or programmed death of cells.
“Our new method using multiplex barcode sequencing technology facilitates the use of pooled shRNA library screening for the identification of combination drug therapy targets,” says Youichi Nakao, member of the research group and professor of biomolecular chemistry at Waseda University.
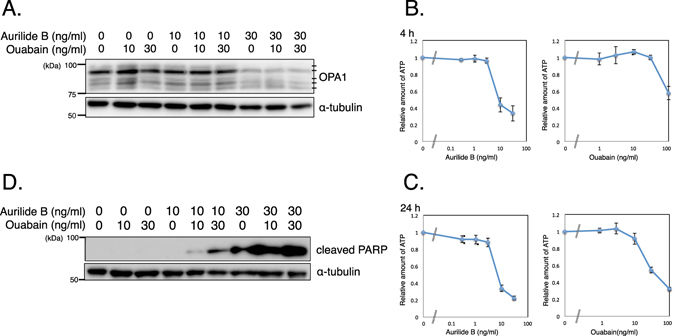
“Combinatorial treatment with aurilide B and oaubain, a toxic substance derived from plants, could trigger apoptosis in specific types of cancer cells with weaker effects to normal cells, suggesting a new kind of treatment.”
This research was published in Scientific Reports.
shRNA in cultured cells can inhibit or “known down” specific genes. shRNA library is a group of various shRNA, and likewise, it creates bioactive compounds with knocked down genes in cell culture. By processing such bioactive compounds, shRNA library screening becomes possible.
“Screening of genome-wide pooled shRNA libraries identifies cellular components that are functionally relevant to the mechanisms of a compound, as well as its direct targets. The Differences between control and compound-treated cell cultures in the growth rates of cells carrying a specific shRNA can be detected by deep sequencing, and this approach has a broader dynamic range than DNA micro arrays.” However, the disadvantage of this type of screening is that it is costly and has low throughput.
To address this issue, the research team used multiplex barcode sequencing technology to conduct a parallel analysis of multiple samples by adding sample-specific index tags to polymerase chain reaction (PCR) primers during sequence library preparation.
“In this study, we introduced a strategy for increasing the throughput of shRNA identification after screening with pooled and barcoded shRNA libraries,” explains Professor Nakao. “The addition of a sample-specific index tag to a PCR primer during sequence library preparation enabled sorting of samples analyzed in parallel by deep sequencing. We show that multiplex barcode sequencing is a reproducible, quantitative, and feasible approach for identifying molecular targets of bioactive compounds.”
“We then used our method to identify genes that modify cellular sensitivity to aurilide B. The results revealed ATP1A1, which encodes a catalytic α-subunit of Na+/K+ ATPase, as a gene involved in survival of cells treated with aurilide B. Indeed, ouabain, a known inhibitor that binds to α-subunits of Na+/K+ ATPase, potentiated the cytotoxicity of aurilide B.”
This study shows that genome-wide shRNA library screening can identify unexpected indirect target genes of bioactive compounds and that it is a novel mechanism underlying aurilide B-induced cancer cell death.
Reference
- Published in: Scientific Reports
- Title: A quantitative shRNA screen identifies ATP1A1 as a gene that regulates cytotoxicity by aurilide B
- Authors: Shohei Takase, Rumi Kurokawa, Daisuke Arai, Kind Kanemoto Kanto, Tatsufumi Okino, Yoichi Nakao, Tetsuo Kushiro, Minoru Yoshida, and Ken Matsumoto
- DOI:10.1038/s41598-017-02016-4